SFB 80:
RNA-DECO
Decorating RNA
for a purpose
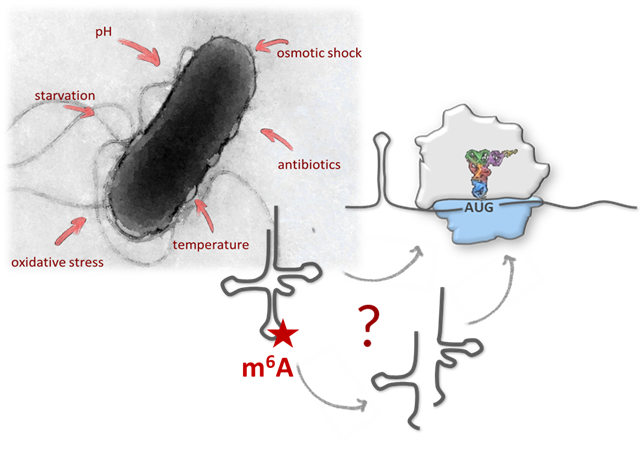
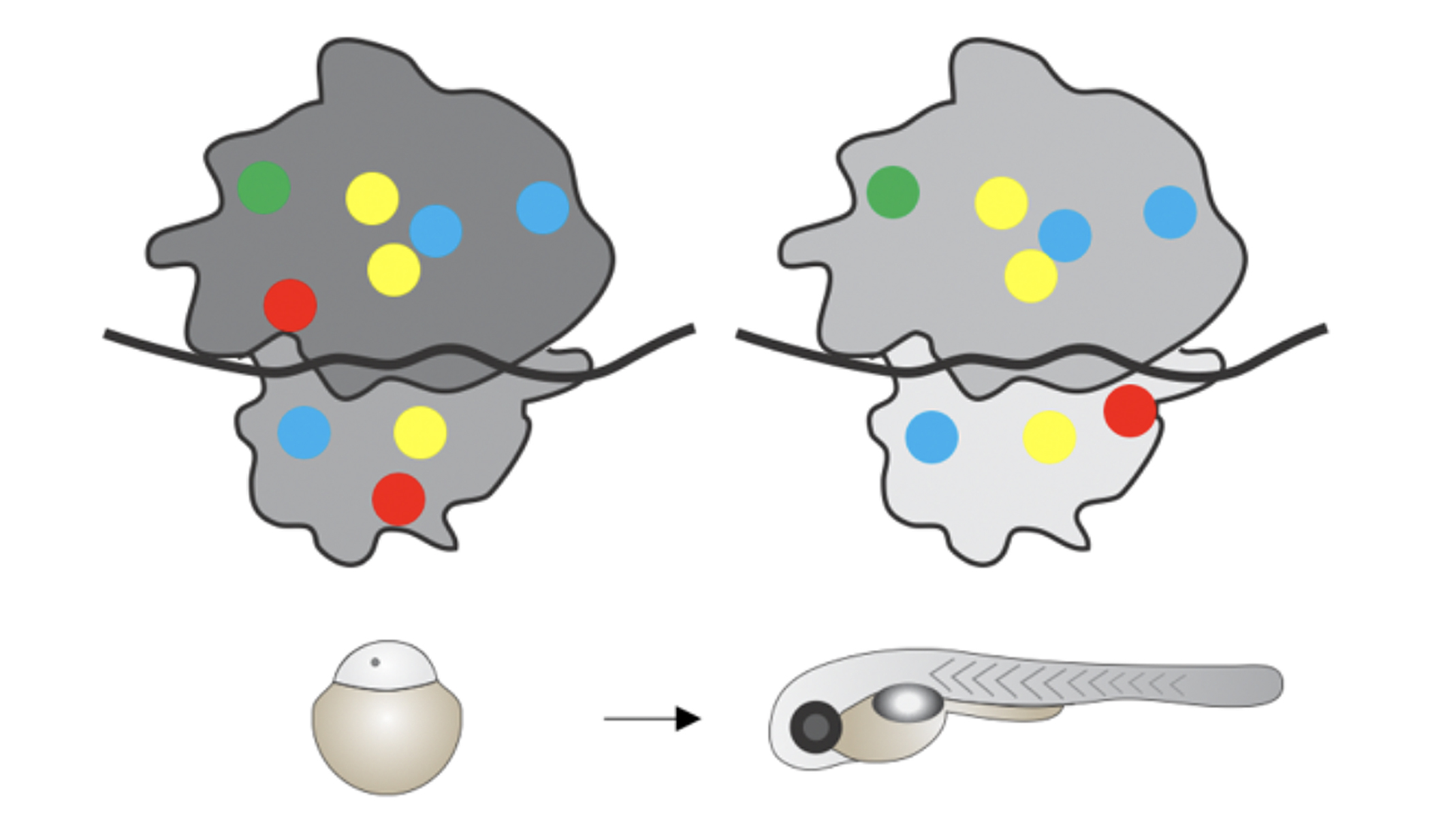
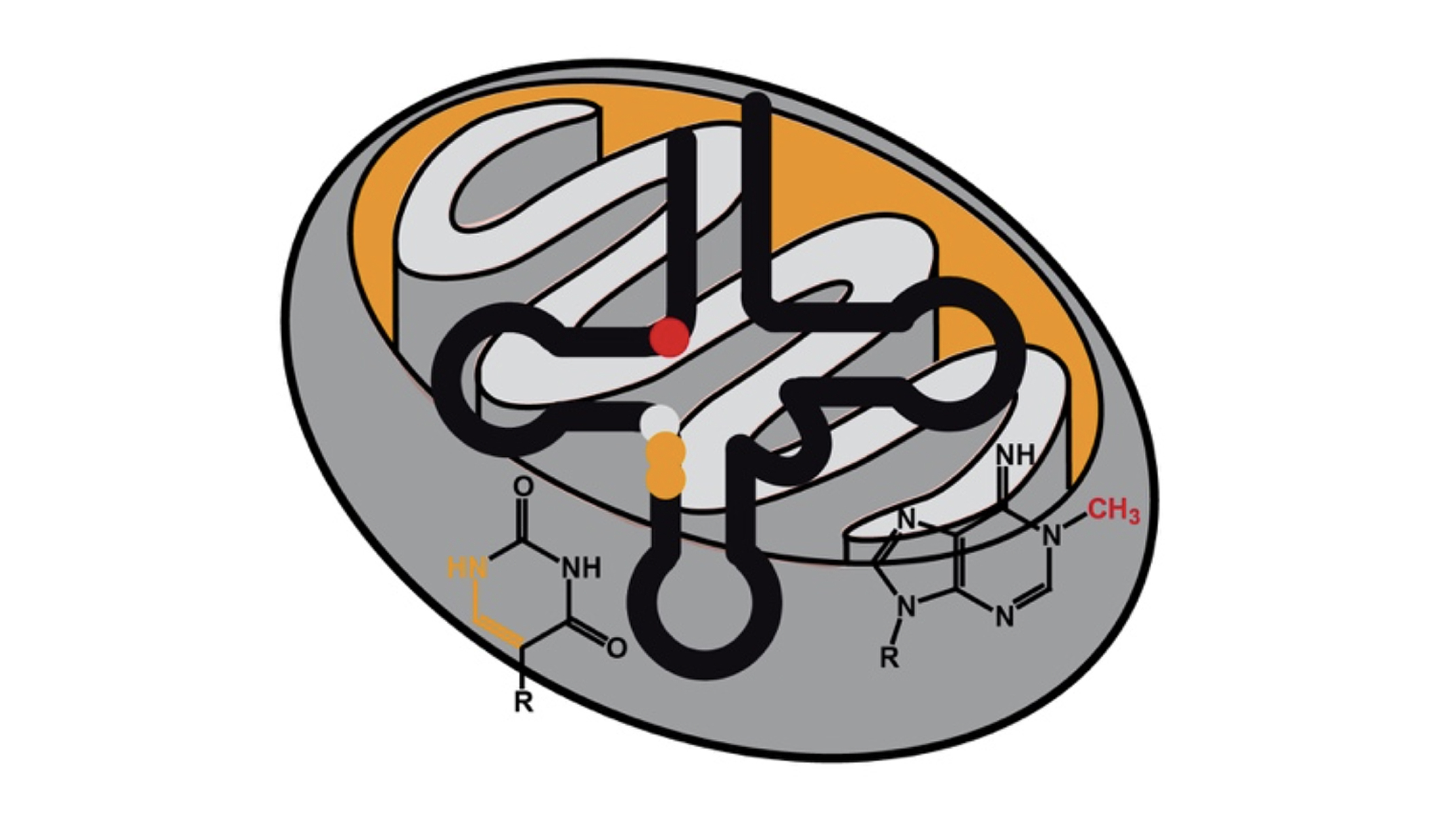
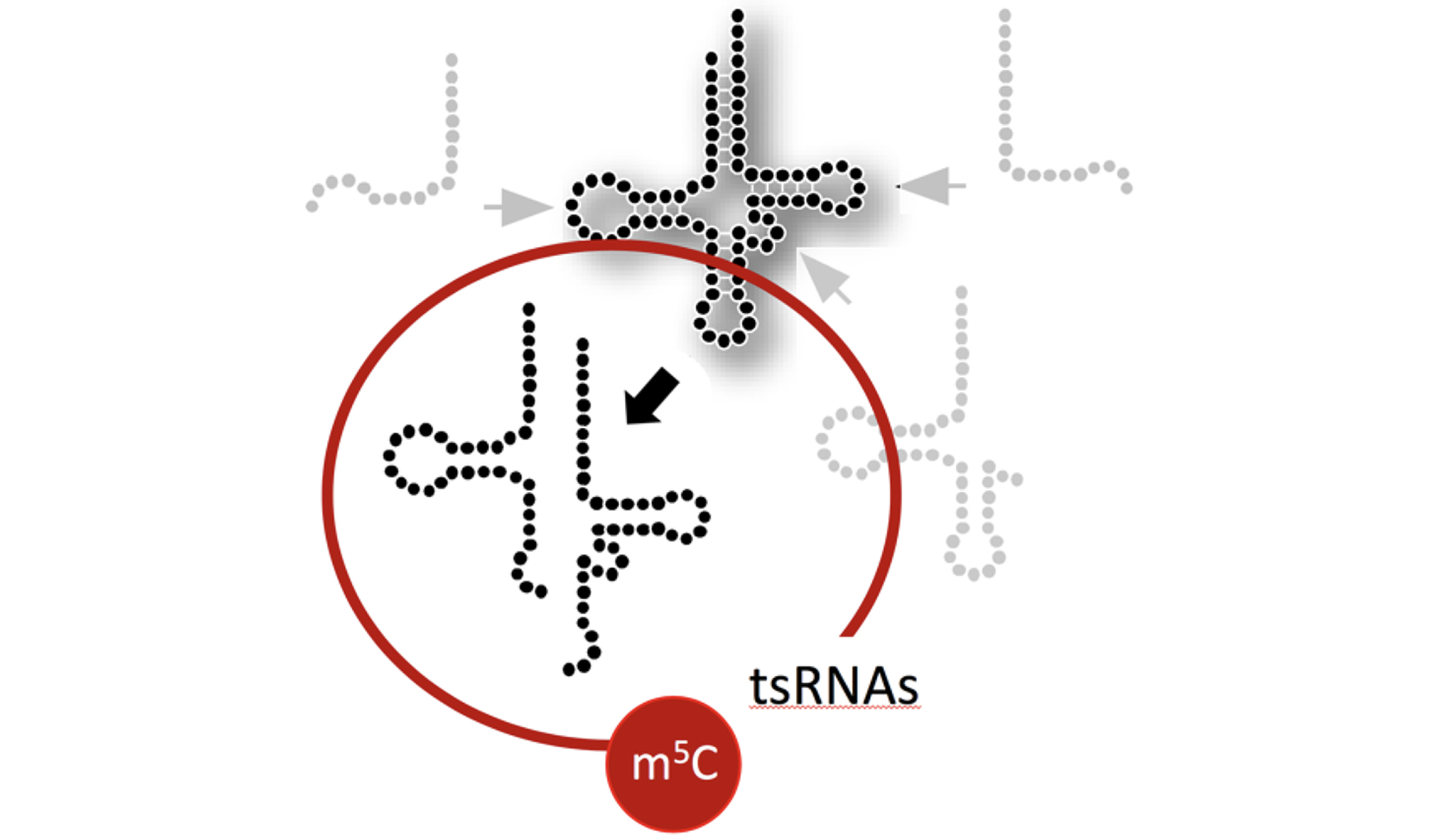
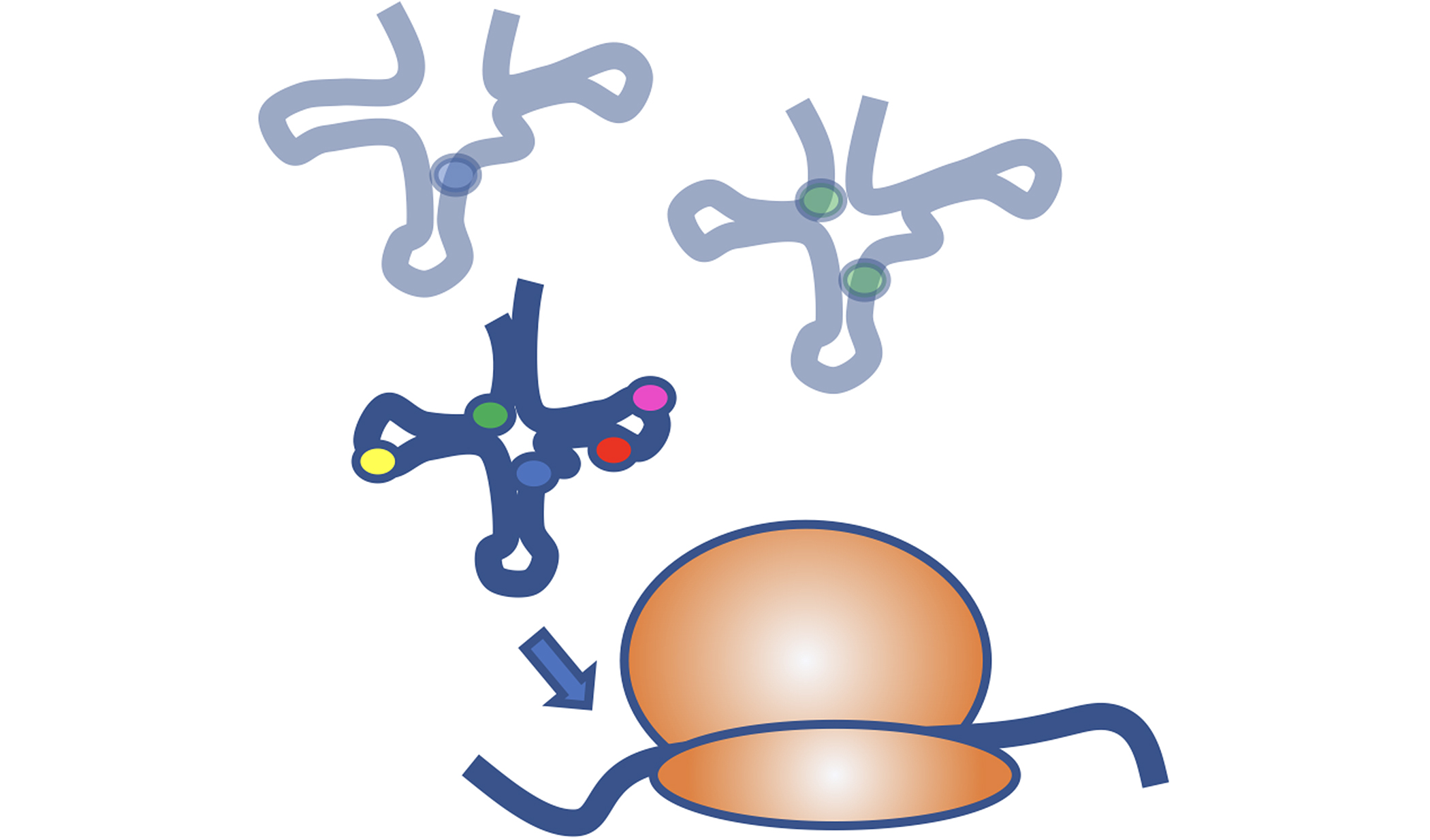
Chemical modifications of RNAs allow the dynamic adaptation of organisms to respond to developmental or environmental changes.
Mission
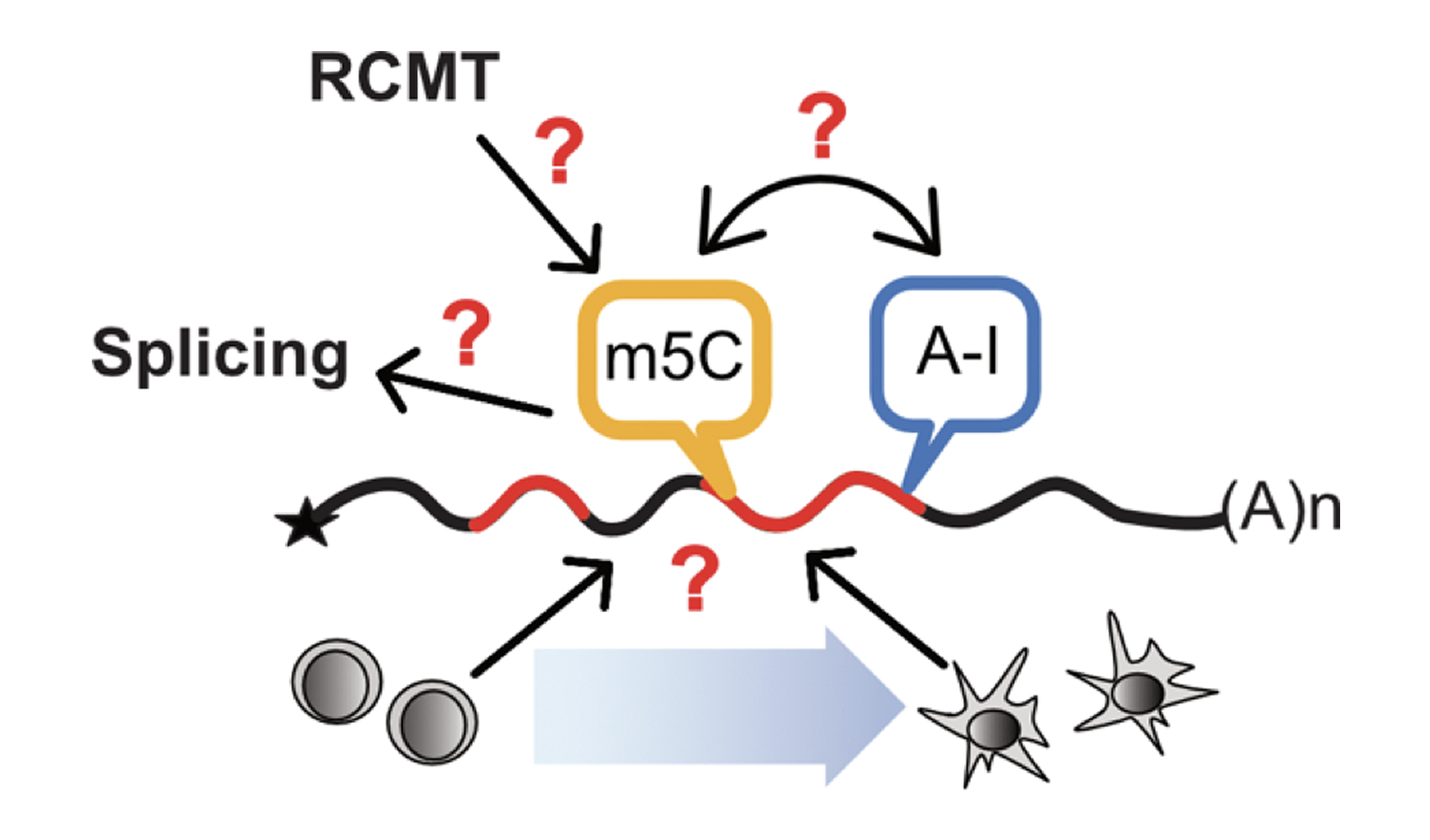
At SFB (F80), RNA-DECO 12 research groups in Austria comprising of biochemists, biologists and bioinformaticians jointly aim at understanding the impact of chemical modifications on:
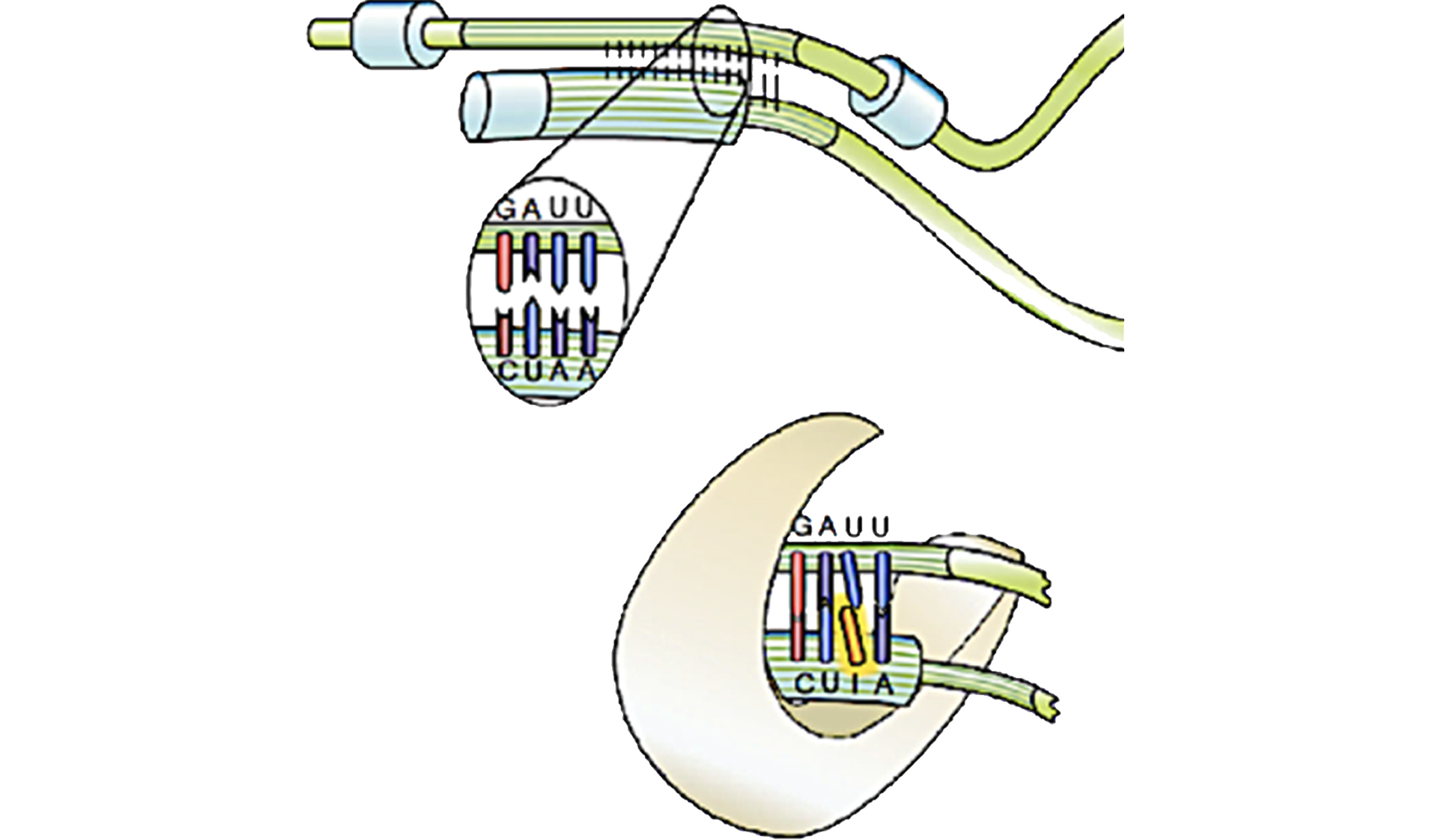
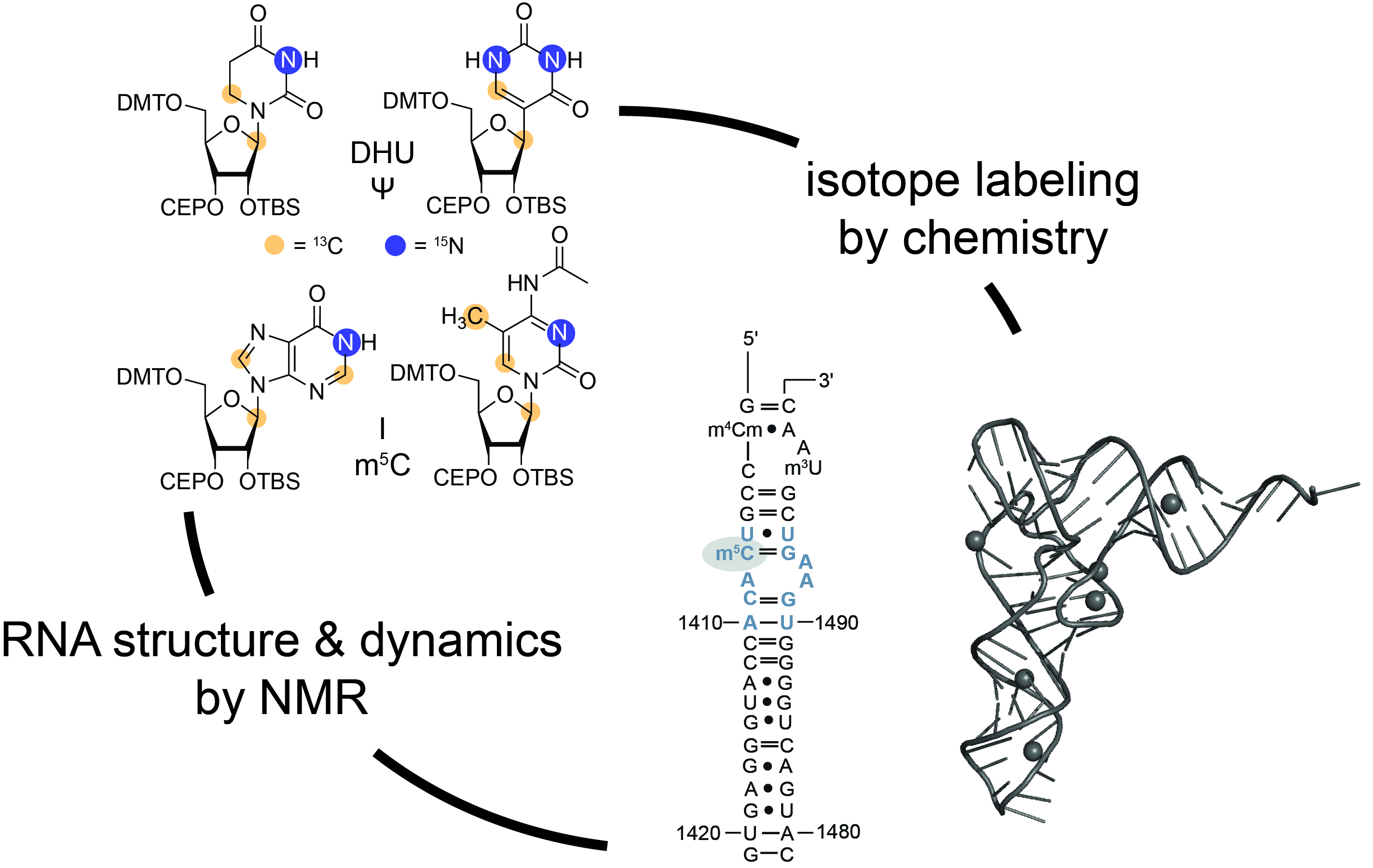
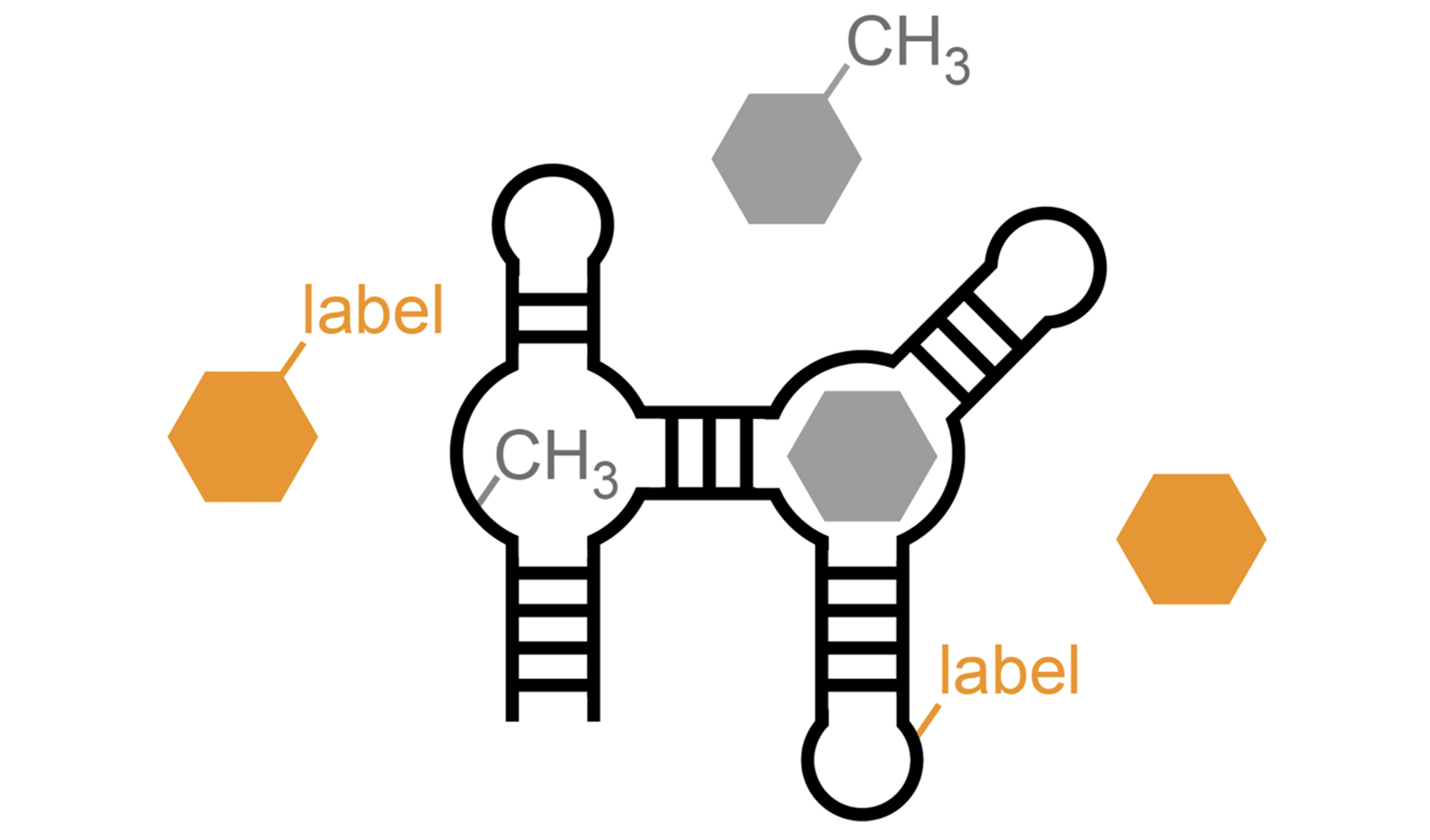
Structure & Function of specific RNAs using different models.
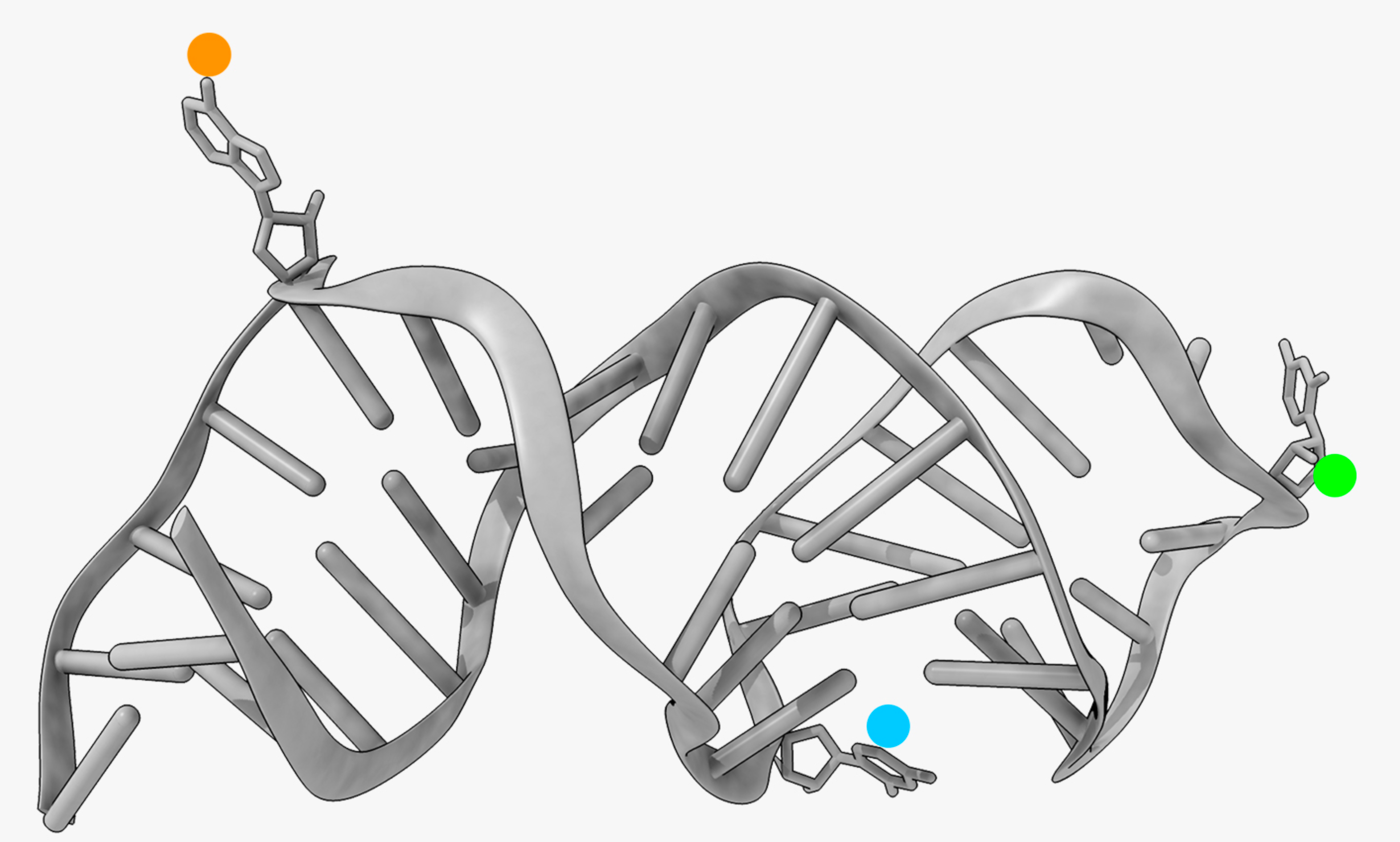
RNA modifications affect

Participants
Scientific Advisory Board
News
Article about importance of RNA in New York times
Nobel Laureate Tom Cech describes importance of RNA and RNA modifications in a New York times article.
Maria Kompatscher defended her Ph.D. thesis successfully!
Maria Kompatscher from Matthias Erlacher’s lab successfully defended her Ph.D. thesis on “Effects of tRNA sequence and modifications on various aspects of translation”.
MID-TERM MEETING - The first four years
The mid-term meeting of RNA DECO was organized at the CCB in Innsbruck February 1st and 2nd.

Publications
tRNA expression and modification landscapes, and their dynamics during zebrafish embryo development
Tom Rappol, Maria Waldl, Anastasia Chugunova, Ivo L Hofacker, Andrea Pauli, Elisa Vilardo
Nucleic Acids Research gkae595 (2024)
Proteome-scale tagging and functional screening in mammalian cells by ORFtag
Filip Nemčko, Moritz Himmelsbach, Vincent Loubiere, Ramesh Yelagandula, Michaela Pagani, Nina Fasching, Julius Brennecke, Ulrich Elling, Alexander Stark, Stefan L. Ameres
Nature Methods (2024)
Splice_sim: a nucleotide conversion-enabled RNA-seq simulation and evaluation framework
Niko Popitsch, Tobias Neumann, Arndt von Haeseler, Stefan L. Ameres
Genome Biology 25 (166) (2024)
Open Positions
Research technician position (40 h) • open since July 23 2024
Research technician position in RNA modification biology
Medical University of Vienna
Center for Anatomy and Cell Biology
Elisa Vilardo, Ph.D.
Schwarzspanierstr. 17, A-1090 Vienna
Tel.: +43 1 40160 37724
E-mail: elisa.vilardo@meduniwien.ac.at
Group Vilardo
The group of Elisa Vilardo at the Center for Anatomy and Cell Biology is recruiting a Technical Assistant. The group works on the biology of tRNA modification, its impact on cell physiology and its implication in human disease. The projects involve the use of a diverse array of techniques, including tissue culture, molecular biology and biochemistry. We offer a young and international working environment within the frame of the Medical University of Vienna, located at the very heart of the city.
The ideal candidate will have a degree in biology, biochemistry or related fields, and will be highly motivated to perform research in the life-sciences with the highest standards. Previous experience in cell and molecular biology is welcome, although further training will be provided in house as needed for the projects. The working language at the institute is English; knowledge of German will be of advantage.
The perspective start date is October 1st 2024. We will be reviewing applications on the go and will keep the job open until the position is filled.
If you are interested, please send a cover letter, CV, and two reference letters to elisa.vilardo@meduniwien.ac.at
Vilardo Tech ad
Open PhD Position • open since April 05 2024
Open PhD Position at the Institute of Genomics and RNomics, Medical University of Innsbruck, Austria
Medical University of Innsbruck
Institute of Genomics and RNomics
Prof. Dr. Matthias Erlacher
Innrain 80, 6020 Innsbruck
Email: Matthias.Erlacher@i-med.ac.at
Erlacher lab
RNA modifications as regulatory elements for protein synthesis
A fully funded PhD position (30 hours per week) is available in the research group of Matthias Erlacher, at the Institute of Genomics and RNomics at the Medical University of Innsbruck, Austria.
Our research group aims to better understand the process of protein synthesis in bacteria and in eukaryotes. In particular, we focus on how modifications of RNA nucleotides can shape specific steps of translation. mRNAs, rRNA and tRNAs, which are the main players in translation, are decorated by a variety of RNA modifications and have been shown to be essential for all organisms. In this project, we aim to deepen our understanding of the regulatory and functional role of certain tRNA and mRNAs modifications.
The available PhD position is embedded in the SFB RNA-deco, which is a consortium of several research teams from Vienna and Innsbruck that study different aspects of RNA modifications.
We are seeking for a highly motivated PhD student with an interest in RNA biology and protein synthesis.
Your Profile:
• Master’s degree in biology, molecular biology, biochemistry or similar
• Interest in basic research on bacterial an eukaryotic translation systems
• Team player: we are a small group so we support/help each other
We offer:
• Highly motivated team and supportive environment
• Possibility to attend national and international conferences and retreats
• Fully funded position available from May 2024; salary according to FWF
The Medical University is located in Innsbruck in the heart of the alps. The city provides numerous possibilities for outdoor activities, such as skiing, hiking or biking. Innsbruck is also home of two large universities and therefore daily life is strongly shaped by students.
If you are interested, please e-mail a CV and letter of motivation to: Matthias.Erlacher@i-med.ac.at
Postdoctoral Position • open since February 12 2024
Postdoctoral Position Moll Lab
Prof. Isabella Mol
MAX PERUTZ LABS
UNIVERSITY OF VIENNA
Vienna BioCenter (VBC)
Dr.-Bohr-Gasse 9
1030 Vienna
Isabella.moll@univie.ac.at
https://www.maxperutzlabs.ac.at/moll
About the Moll lab
Our group aims to unravel molecular mechanisms underlying the post-transcriptional regulation of gene expression in Bacteria, with a special focus on the regulatory capacity of the ribosome. We investigate the intricacies of ribosome heterogeneity and reprogramming of protein synthesis in vivo and in vitro, utilizing a comprehensive methodology that integrates advanced techniques in molecular biology and bacterial physi-ology employing the well-established model organism Escherichia coli. Additionally, we enhance our research with structural studies to gain further insights into the various mechanisms that control ribosome diversity and selectivity. Current projects are aimed at understanding the role of conditional RNA modifications on translation dynamics in the bacterial stress response, which are embedded in the special research program SFB RNA-DECO (https://www.rna-deco.org/), representing a consortium of top RNA research labs in Austria investigating RNA modifications.
We are looking for a skilled, enthusiastic, and motivated postdoctoral scientist to join our team. The success-ful applicant will take a lead role in the project on regulatory roles of tRNA modifications in protein synthesis. Training and supervision will be provided throughout the project, but we also expect a high level of drive and independence. As part of our group, you will closely collaborate with colleagues and the group leader and contribute to the development of the team. We strive for a supportive feedback culture, a diverse and inter-national team, and a place for creative and rigorous science.
This is part of your personality:
- PhD degree in Microbiology, Molecular Biology, Biochemistry, or related fields.
- Research competence and initiative proven through international publications in peer-reviewed journals
- Extensive experience in molecular microbiology with a special emphasis on RNA biochemistry, tran-scriptomics, especially ribosome profiling, and bioinformatic data analysis. Previous experience with structural biology is a plus.
- Excellent command of written and spoken English.
- Team player and high social/communicative skills.
Your future tasks:
You actively participate in research, teaching, and administration, which means:
- Contribution to the project on regulatory roles of tRNA modifications in protein synthesis.
- Participation in writing scientific publications and grant applications.
- Participation in teaching and supervision of BSc and MSc students.
- You participate in the organization of meetings, conferences, and symposia.
What we offer:
Employment duration: The position is limited to 3 years and offers the possibility of extension for 1 year.
Fair salary: Job grading in accordance with collective bargaining agreement. The basic salary of EUR 4,351.90 (14 times a year) increases if we can credit professional experience.
Work-life balance: Our employees enjoy flexible, family-friendly working hours.
Inspiring working atmosphere: You are a part of an international academic team in a healthy and fair work-ing environment.
Good public transport connections: Your workplace in the centre of beautiful Vienna is easily accessible by public transport.
Internal further training & Coaching: Opportunity to deepen your skills on an ongoing basis.
Equal opportunities for everyone: We look forward to diverse personalities in the team!
It is that easy to apply:
Interested applicants should send their CV, a motivation letter (describing experience and research interests), as well as the contact information for at least two references via e-mail with subject line ‘Postdoc applica-tion’ to Isabella Moll (Isabella.moll@univie.ac.at).
The position is available immediately with a flexible start date.
About the Max Perutz Labs
The Max Perutz Labs are a research institute established by the University of Vienna and the Medical Univer-sity of Vienna to provide an environment for excellent, internationally recognized research and education in the field of Molecular Biology. Dedicated to a mechanistic understanding of fundamental biomedical process-es,
scientists at the Max Perutz Labs aim to link breakthroughs in basic research to advances in human health. The Max Perutz Labs are located at the Vienna BioCenter, one of Europe’s hotspots for Life Sciences, and host 45 research groups, involving around 400 scientists and staff from more than 50 nations.
www.maxperutzlabs.ac.at